Use the Pandas Dataframe Dept stats Again in Python
Watch Now This tutorial has a related video course created by the Real Python team. Picket it together with the written tutorial to deepen your agreement: The Pandas DataFrame: Working With Information Efficiently
The Pandas DataFrame is a structure that contains 2-dimensional data and its corresponding labels. DataFrames are widely used in information science, machine learning, scientific computing, and many other data-intensive fields.
DataFrames are similar to SQL tables or the spreadsheets that you lot work with in Excel or Calc. In many cases, DataFrames are faster, easier to use, and more than powerful than tables or spreadsheets because they're an integral role of the Python and NumPy ecosystems.
In this tutorial, you lot'll learn:
- What a Pandas DataFrame is and how to create ane
- How to access, alter, add, sort, filter, and delete data
- How to handle missing values
- How to work with time-series data
- How to chop-chop visualize data
Information technology's time to get started with Pandas DataFrames!
Introducing the Pandas DataFrame
Pandas DataFrames are data structures that contain:
- Data organized in ii dimensions, rows and columns
- Labels that stand for to the rows and columns
You can start working with DataFrames past importing Pandas:
>>>
>>> import pandas equally pd At present that you have Pandas imported, you lot can work with DataFrames.
Imagine you're using Pandas to analyze data about job candidates for a position developing spider web applications with Python. Say you're interested in the candidates' names, cities, ages, and scores on a Python programming examination, or py-score:
name | city | historic period | py-score | |
|---|---|---|---|---|
101 | Xavier | Mexico City | 41 | 88.0 |
102 | Ann | Toronto | 28 | 79.0 |
103 | Jana | Prague | 33 | 81.0 |
104 | Yi | Shanghai | 34 | eighty.0 |
105 | Robin | Manchester | 38 | 68.0 |
106 | Amal | Cairo | 31 | 61.0 |
107 | Nori | Osaka | 37 | 84.0 |
In this table, the first row contains the column labels (name, urban center, age, and py-score). The first cavalcade holds the row labels (101, 102, and so on). All other cells are filled with the data values.
Now you lot accept everything you demand to create a Pandas DataFrame.
There are several ways to create a Pandas DataFrame. In most cases, you'll use the DataFrame constructor and provide the data, labels, and other data. You can pass the information equally a two-dimensional list, tuple, or NumPy assortment. You can also pass it every bit a dictionary or Pandas Series instance, or as one of several other data types not covered in this tutorial.
For this case, assume you lot're using a dictionary to pass the information:
>>>
>>> data = { ... 'proper noun' : [ 'Xavier' , 'Ann' , 'Jana' , 'Yi' , 'Robin' , 'Amal' , 'Nori' ], ... 'urban center' : [ 'United mexican states City' , 'Toronto' , 'Prague' , 'Shanghai' , ... 'Manchester' , 'Cairo' , 'Osaka' ], ... 'age' : [ 41 , 28 , 33 , 34 , 38 , 31 , 37 ], ... 'py-score' : [ 88.0 , 79.0 , 81.0 , 80.0 , 68.0 , 61.0 , 84.0 ] ... } >>> row_labels = [ 101 , 102 , 103 , 104 , 105 , 106 , 107 ] data is a Python variable that refers to the dictionary that holds your candidate data. It too contains the labels of the columns:
-
'proper name' -
'city' -
'age' -
'py-score'
Finally, row_labels refers to a list that contains the labels of the rows, which are numbers ranging from 101 to 107.
Now you're set to create a Pandas DataFrame:
>>>
>>> df = pd . DataFrame ( data = data , index = row_labels ) >>> df name city age py-score 101 Xavier Mexico City 41 88.0 102 Ann Toronto 28 79.0 103 Jana Prague 33 81.0 104 Yi Shanghai 34 eighty.0 105 Robin Manchester 38 68.0 106 Amal Cairo 31 61.0 107 Nori Osaka 37 84.0 That's it! df is a variable that holds the reference to your Pandas DataFrame. This Pandas DataFrame looks just similar the candidate table in a higher place and has the following features:
- Row labels from
101to107 - Column labels such every bit
'proper noun','metropolis','historic period', and'py-score' - Data such equally candidate names, cities, ages, and Python test scores
This effigy shows the labels and information from df:
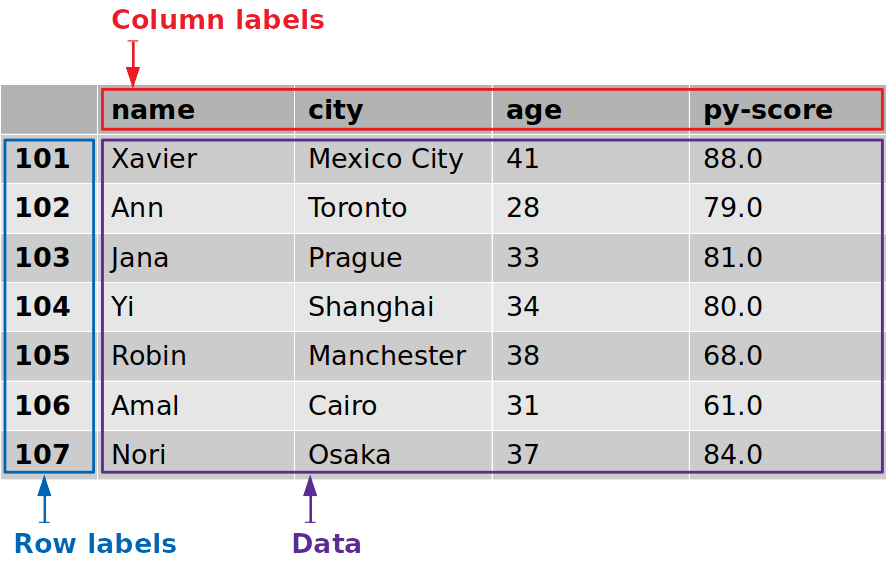
The row labels are outlined in blue, whereas the column labels are outlined in red, and the information values are outlined in purple.
Pandas DataFrames tin can sometimes exist very big, making it impractical to look at all the rows at once. You tin apply .head() to show the first few items and .tail() to bear witness the terminal few items:
>>>
>>> df . head ( north = 2 ) proper noun urban center age py-score 101 Xavier Mexico Urban center 41 88.0 102 Ann Toronto 28 79.0 >>> df . tail ( due north = ii ) name city historic period py-score 106 Amal Cairo 31 61.0 107 Nori Osaka 37 84.0 That'south how you can testify only the outset or terminate of a Pandas DataFrame. The parameter northward specifies the number of rows to show.
Y'all can admission a column in a Pandas DataFrame the same way you would get a value from a lexicon:
>>>
>>> cities = df [ 'urban center' ] >>> cities 101 Mexico Urban center 102 Toronto 103 Prague 104 Shanghai 105 Manchester 106 Cairo 107 Osaka Name: urban center, dtype: object This is the near convenient way to get a column from a Pandas DataFrame.
If the proper name of the column is a string that is a valid Python identifier, and so you can use dot notation to access information technology. That is, yous can access the cavalcade the same way you would get the attribute of a form instance:
>>>
>>> df . city 101 Mexico City 102 Toronto 103 Prague 104 Shanghai 105 Manchester 106 Cairo 107 Osaka Name: city, dtype: object That's how y'all get a particular column. Y'all've extracted the column that corresponds with the label 'city', which contains the locations of all your job candidates.
It'southward important to notice that you've extracted both the information and the corresponding row labels:
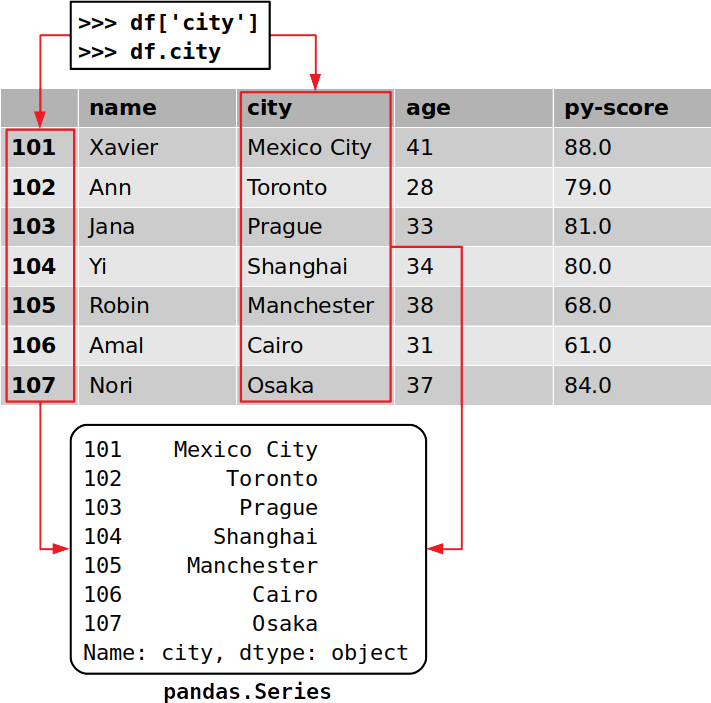
Each cavalcade of a Pandas DataFrame is an instance of pandas.Serial, a structure that holds ane-dimensional information and their labels. You can become a single particular of a Series object the same fashion yous would with a lexicon, by using its label as a key:
>>>
>>> cities [ 102 ] 'Toronto' In this example, 'Toronto' is the data value and 102 is the corresponding characterization. Equally you'll see in a later section, there are other ways to get a particular item in a Pandas DataFrame.
You tin also access a whole row with the accessor .loc[]:
>>>
>>> df . loc [ 103 ] name Jana urban center Prague age 33 py-score 81 Proper name: 103, dtype: object This time, yous've extracted the row that corresponds to the label 103, which contains the data for the candidate named Jana. In addition to the data values from this row, y'all've extracted the labels of the respective columns:
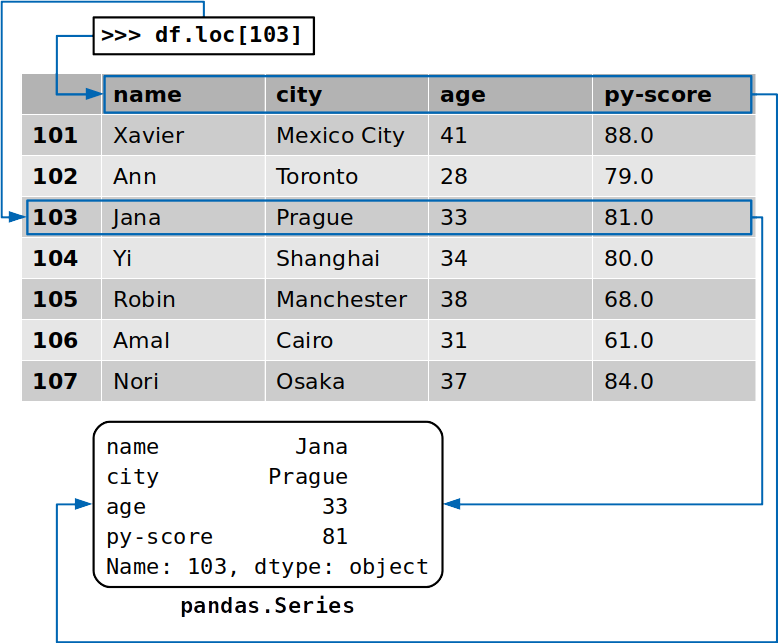
The returned row is also an case of pandas.Serial.
Creating a Pandas DataFrame
Equally already mentioned, there are several way to create a Pandas DataFrame. In this section, you lot'll larn to do this using the DataFrame constructor along with:
- Python dictionaries
- Python lists
- Two-dimensional NumPy arrays
- Files
There are other methods as well, which you can learn about in the official documentation.
You can kickoff by importing Pandas along with NumPy, which you'll use throughout the following examples:
>>>
>>> import numpy as np >>> import pandas equally pd That'southward information technology. Now you're set up to create some DataFrames.
Creating a Pandas DataFrame With Dictionaries
As you've already seen, y'all can create a Pandas DataFrame with a Python dictionary:
>>>
>>> d = { 'x' : [ one , two , 3 ], 'y' : np . array ([ 2 , 4 , 8 ]), 'z' : 100 } >>> pd . DataFrame ( d ) x y z 0 one two 100 1 2 4 100 2 3 8 100 The keys of the lexicon are the DataFrame's cavalcade labels, and the dictionary values are the data values in the corresponding DataFrame columns. The values can be contained in a tuple, list, one-dimensional NumPy array, Pandas Series object, or i of several other information types. You can too provide a unmarried value that volition be copied along the entire column.
It's possible to control the order of the columns with the columns parameter and the row labels with index:
>>>
>>> pd . DataFrame ( d , index = [ 100 , 200 , 300 ], columns = [ 'z' , 'y' , '10' ]) z y x 100 100 2 i 200 100 iv two 300 100 8 3 As yous tin come across, you've specified the row labels 100, 200, and 300. You've likewise forced the order of columns: z, y, x.
Creating a Pandas DataFrame With Lists
Another way to create a Pandas DataFrame is to use a listing of dictionaries:
>>>
>>> l = [{ 'x' : 1 , 'y' : 2 , 'z' : 100 }, ... { '10' : two , 'y' : 4 , 'z' : 100 }, ... { 'x' : 3 , 'y' : eight , 'z' : 100 }] >>> pd . DataFrame ( l ) x y z 0 1 2 100 i 2 4 100 2 3 eight 100 Over again, the dictionary keys are the cavalcade labels, and the dictionary values are the information values in the DataFrame.
You can also apply a nested list, or a list of lists, as the data values. If yous do, then it's wise to explicitly specify the labels of columns, rows, or both when y'all create the DataFrame:
>>>
>>> l = [[ 1 , ii , 100 ], ... [ 2 , iv , 100 ], ... [ 3 , eight , 100 ]] >>> pd . DataFrame ( fifty , columns = [ 'x' , 'y' , 'z' ]) ten y z 0 i 2 100 i 2 four 100 2 3 viii 100 That'south how you tin can apply a nested list to create a Pandas DataFrame. You can likewise use a list of tuples in the aforementioned way. To practice then, just supersede the nested lists in the example in a higher place with tuples.
Creating a Pandas DataFrame With NumPy Arrays
You can pass a two-dimensional NumPy array to the DataFrame constructor the same style you do with a listing:
>>>
>>> arr = np . assortment ([[ one , two , 100 ], ... [ 2 , iv , 100 ], ... [ 3 , 8 , 100 ]]) >>> df_ = pd . DataFrame ( arr , columns = [ 'ten' , 'y' , 'z' ]) >>> df_ x y z 0 ane ii 100 1 2 four 100 two 3 8 100 Although this example looks almost the same equally the nested listing implementation above, it has one advantage: You can specify the optional parameter copy.
When copy is fix to False (its default setting), the data from the NumPy array isn't copied. This means that the original information from the array is assigned to the Pandas DataFrame. If you modify the array, then your DataFrame will alter as well:
>>>
>>> arr [ 0 , 0 ] = thousand >>> df_ x y z 0 grand two 100 1 2 4 100 2 3 8 100 As you can run across, when you modify the first item of arr, you besides modify df_.
If this behavior isn't what you lot want, and so you should specify re-create=Truthful in the DataFrame constructor. That mode, df_ will exist created with a copy of the values from arr instead of the bodily values.
Creating a Pandas DataFrame From Files
Yous can relieve and load the data and labels from a Pandas DataFrame to and from a number of file types, including CSV, Excel, SQL, JSON, and more. This is a very powerful characteristic.
Yous can save your chore candidate DataFrame to a CSV file with .to_csv():
>>>
>>> df . to_csv ( 'data.csv' ) The statement higher up will produce a CSV file called data.csv in your working directory:
,name,city,historic period,py-score 101,Xavier,Mexico Metropolis,41,88.0 102,Ann,Toronto,28,79.0 103,Jana,Prague,33,81.0 104,Yi,Shanghai,34,lxxx.0 105,Robin,Manchester,38,68.0 106,Amal,Cairo,31,61.0 107,Nori,Osaka,37,84.0 Now that you have a CSV file with data, yous can load it with read_csv():
>>>
>>> pd . read_csv ( 'data.csv' , index_col = 0 ) name urban center historic period py-score 101 Xavier Mexico City 41 88.0 102 Ann Toronto 28 79.0 103 Jana Prague 33 81.0 104 Yi Shanghai 34 eighty.0 105 Robin Manchester 38 68.0 106 Amal Cairo 31 61.0 107 Nori Osaka 37 84.0 That'due south how you get a Pandas DataFrame from a file. In this case, index_col=0 specifies that the row labels are located in the get-go cavalcade of the CSV file.
Retrieving Labels and Data
Now that yous've created your DataFrame, you lot can outset retrieving information from information technology. With Pandas, y'all can perform the following deportment:
- Retrieve and alter row and column labels as sequences
- Correspond data every bit NumPy arrays
- Cheque and adjust the information types
- Analyze the size of
DataFrameobjects
Pandas DataFrame Labels as Sequences
You can become the DataFrame'southward row labels with .index and its column labels with .columns:
>>>
>>> df . index Int64Index([1, 2, 3, iv, 5, half-dozen, 7], dtype='int64') >>> df . columns Alphabetize(['name', 'city', 'age', 'py-score'], dtype='object') At present you have the row and cavalcade labels as special kinds of sequences. Equally you lot tin can with any other Python sequence, you can get a single item:
>>>
>>> df . columns [ one ] 'metropolis' In addition to extracting a particular detail, y'all can utilize other sequence operations, including iterating through the labels of rows or columns. However, this is rarely necessary since Pandas offers other ways to iterate over DataFrames, which yous'll see in a after section.
Yous can also use this approach to modify the labels:
>>>
>>> df . index = np . arange ( 10 , 17 ) >>> df . index Int64Index([10, 11, 12, thirteen, xiv, xv, sixteen], dtype='int64') >>> df proper name urban center age py-score 10 Xavier United mexican states City 41 88.0 11 Ann Toronto 28 79.0 12 Jana Prague 33 81.0 13 Yi Shanghai 34 80.0 14 Robin Manchester 38 68.0 xv Amal Cairo 31 61.0 sixteen Nori Osaka 37 84.0 In this case, yous utilise numpy.arange() to generate a new sequence of row labels that holds the integers from 10 to xvi. To learn more about arange(), check out NumPy arange(): How to Utilise np.arange().
Keep in heed that if you try to modify a particular particular of .index or .columns, then you'll become a TypeError.
Information as NumPy Arrays
Sometimes yous might want to extract data from a Pandas DataFrame without its labels. To get a NumPy assortment with the unlabeled information, you tin utilize either .to_numpy() or .values:
>>>
>>> df . to_numpy () array([['Xavier', 'Mexico Urban center', 41, 88.0], ['Ann', 'Toronto', 28, 79.0], ['Jana', 'Prague', 33, 81.0], ['Yi', 'Shanghai', 34, 80.0], ['Robin', 'Manchester', 38, 68.0], ['Amal', 'Cairo', 31, 61.0], ['Nori', 'Osaka', 37, 84.0]], dtype=object) Both .to_numpy() and .values work similarly, and they both return a NumPy array with the information from the Pandas DataFrame:
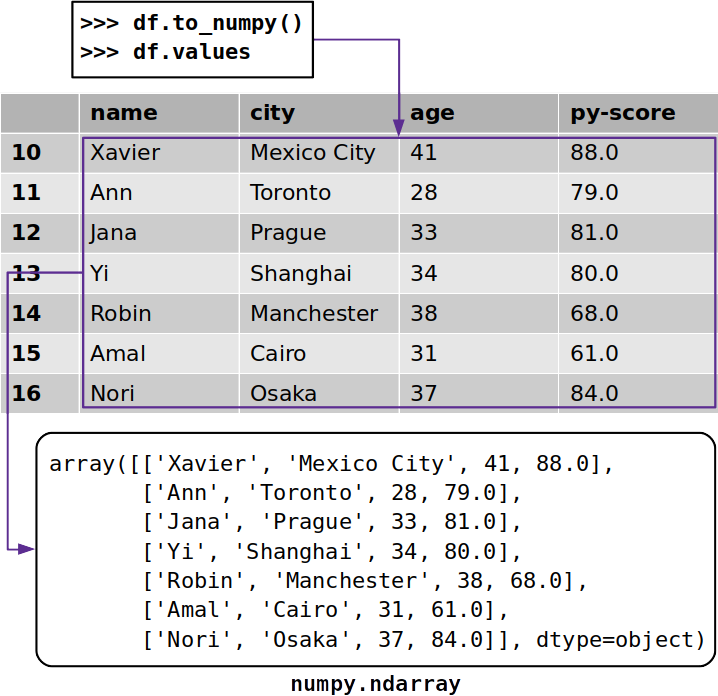
The Pandas documentation suggests using .to_numpy() because of the flexibility offered by 2 optional parameters:
-
dtype: Use this parameter to specify the data type of the resulting assortment. It'south set toNoneby default. -
copy: Set this parameter toFalseif you desire to use the original information from the DataFrame. Set information technology toTrueif you want to make a copy of the information.
Still, .values has been around for much longer than .to_numpy(), which was introduced in Pandas version 0.24.0. That means y'all'll probably see .values more oft, specially in older code.
Data Types
The types of the information values, also called information types or dtypes, are important because they determine the amount of retentivity your DataFrame uses, as well as its calculation speed and level of precision.
Pandas relies heavily on NumPy data types. However, Pandas one.0 introduced some additional types:
-
BooleanDtypeandBooleanArrayback up missing Boolean values and Kleene three-value logic. -
StringDtypeandStringArraystand for a dedicated string type.
You lot can get the data types for each cavalcade of a Pandas DataFrame with .dtypes:
>>>
>>> df . dtypes proper noun object city object age int64 py-score float64 dtype: object As you can see, .dtypes returns a Series object with the column names as labels and the respective information types as values.
If you want to modify the information blazon of one or more columns, then yous can use .astype():
>>>
>>> df_ = df . astype ( dtype = { 'age' : np . int32 , 'py-score' : np . float32 }) >>> df_ . dtypes name object metropolis object historic period int32 py-score float32 dtype: object The most important and but mandatory parameter of .astype() is dtype. Information technology expects a data type or dictionary. If you lot pass a lexicon, then the keys are the column names and the values are your desired corresponding data types.
As you can run into, the information types for the columns age and py-score in the DataFrame df are both int64, which represents 64-flake (or 8-byte) integers. Still, df_ likewise offers a smaller, 32-chip (4-byte) integer information blazon called int32.
Pandas DataFrame Size
The attributes .ndim, .size, and .shape return the number of dimensions, number of information values across each dimension, and total number of data values, respectively:
>>>
>>> df_ . ndim 2 >>> df_ . shape (7, 4) >>> df_ . size 28 DataFrame instances accept two dimensions (rows and columns), so .ndim returns two. A Serial object, on the other mitt, has only a single dimension, then in that case, .ndim would render 1.
The .shape attribute returns a tuple with the number of rows (in this example 7) and the number of columns (4). Finally, .size returns an integer equal to the number of values in the DataFrame (28).
Yous can fifty-fifty cheque the amount of retentivity used by each column with .memory_usage():
>>>
>>> df_ . memory_usage () Alphabetize 56 name 56 metropolis 56 age 28 py-score 28 dtype: int64 As you can see, .memory_usage() returns a Series with the column names as labels and the retention usage in bytes as data values. If yous want to exclude the memory usage of the column that holds the row labels, then pass the optional argument index=False.
In the instance above, the last two columns, age and py-score, use 28 bytes of memory each. That's because these columns take seven values, each of which is an integer that takes 32 $.25, or 4 bytes. 7 integers times 4 bytes each equals a total of 28 bytes of memory usage.
Accessing and Modifying Data
You've already learned how to get a detail row or column of a Pandas DataFrame equally a Series object:
>>>
>>> df [ 'name' ] x Xavier 11 Ann 12 Jana 13 Yi fourteen Robin fifteen Amal xvi Nori Proper noun: name, dtype: object >>> df . loc [ 10 ] proper name Xavier city United mexican states City historic period 41 py-score 88 Proper name: 10, dtype: object In the first example, y'all access the column proper name as yous would access an element from a lexicon, by using its label as a key. If the column label is a valid Python identifier, and then y'all tin can as well employ dot note to access the column. In the second example, you lot use .loc[] to get the row past its characterization, ten.
Getting Information With Accessors
In addition to the accessor .loc[], which you can use to go rows or columns past their labels, Pandas offers the accessor .iloc[], which retrieves a row or cavalcade by its integer index. In well-nigh cases, y'all can utilise either of the two:
>>>
>>> df . loc [ x ] name Xavier city Mexico City historic period 41 py-score 88 Name: 10, dtype: object >>> df . iloc [ 0 ] name Xavier city Mexico City historic period 41 py-score 88 Proper name: 10, dtype: object df.loc[10] returns the row with the label 10. Similarly, df.iloc[0] returns the row with the nix-based index 0, which is the kickoff row. As y'all can run into, both statements return the same row every bit a Series object.
Pandas has iv accessors in total:
-
.loc[]accepts the labels of rows and columns and returns Series or DataFrames. Y'all tin can utilize it to get entire rows or columns, every bit well as their parts. -
.iloc[]accepts the null-based indices of rows and columns and returns Series or DataFrames. Yous can use it to get entire rows or columns, or their parts. -
.at[]accepts the labels of rows and columns and returns a single data value. -
.iat[]accepts the null-based indices of rows and columns and returns a single information value.
Of these, .loc[] and .iloc[] are particularly powerful. They support slicing and NumPy-manner indexing. You can utilise them to admission a column:
>>>
>>> df . loc [:, 'city' ] 10 Mexico City 11 Toronto 12 Prague 13 Shanghai 14 Manchester 15 Cairo 16 Osaka Proper name: city, dtype: object >>> df . iloc [:, 1 ] 10 Mexico Metropolis eleven Toronto 12 Prague 13 Shanghai 14 Manchester 15 Cairo 16 Osaka Proper noun: city, dtype: object df.loc[:, 'city'] returns the column city. The slice construct (:) in the row characterization place ways that all the rows should exist included. df.iloc[:, one] returns the aforementioned cavalcade because the zip-based index 1 refers to the second cavalcade, urban center.
Just as you can with NumPy, yous can provide slices along with lists or arrays instead of indices to get multiple rows or columns:
>>>
>>> df . loc [ 11 : 15 , [ 'name' , 'city' ]] proper noun city 11 Ann Toronto 12 Jana Prague thirteen Yi Shanghai 14 Robin Manchester 15 Amal Cairo >>> df . iloc [ 1 : 6 , [ 0 , one ]] name metropolis eleven Ann Toronto 12 Jana Prague 13 Yi Shanghai 14 Robin Manchester fifteen Amal Cairo In this example, you apply:
- Slices to become the rows with the labels
11through15, which are equivalent to the indices1through5 - Lists to get the columns
nameandcity, which are equivalent to the indices0andi
Both statements render a Pandas DataFrame with the intersection of the desired 5 rows and two columns.
This brings up a very important difference between .loc[] and .iloc[]. As you can run into from the previous example, when y'all pass the row labels 11:15 to .loc[], you become the rows 11 through 15. Still, when you laissez passer the row indices 1:half-dozen to .iloc[], you lot just get the rows with the indices ane through 5.
The reason you lot only get indices 1 through 5 is that, with .iloc[], the stop alphabetize of a slice is exclusive, significant it is excluded from the returned values. This is consistent with Python sequences and NumPy arrays. With .loc[], nevertheless, both start and stop indices are inclusive, pregnant they are included with the returned values.
You can skip rows and columns with .iloc[] the same mode you tin with slicing tuples, lists, and NumPy arrays:
>>>
>>> df . iloc [ ane : 6 : 2 , 0 ] 11 Ann 13 Yi 15 Amal Name: name, dtype: object In this example, you specify the desired row indices with the piece ane:six:2. This means that you commencement with the row that has the index 1 (the second row), terminate before the row with the index six (the seventh row), and skip every second row.
Instead of using the slicing construct, you could also utilize the built-in Python class piece(), too equally numpy.s_[] or pd.IndexSlice[]:
>>>
>>> df . iloc [ slice ( 1 , 6 , 2 ), 0 ] xi Ann xiii Yi fifteen Amal Proper noun: name, dtype: object >>> df . iloc [ np . s_ [ ane : half-dozen : ii ], 0 ] 11 Ann 13 Yi fifteen Amal Name: name, dtype: object >>> df . iloc [ pd . IndexSlice [ ane : half-dozen : 2 ], 0 ] 11 Ann 13 Yi 15 Amal Proper noun: name, dtype: object You might observe 1 of these approaches more convenient than others depending on your situation.
Information technology's possible to utilize .loc[] and .iloc[] to get particular data values. However, when you lot demand only a unmarried value, Pandas recommends using the specialized accessors .at[] and .iat[]:
>>>
>>> df . at [ 12 , 'name' ] 'Jana' >>> df . iat [ ii , 0 ] 'Jana' Here, yous used .at[] to get the name of a single candidate using its respective column and row labels. You likewise used .iat[] to retrieve the aforementioned name using its column and row indices.
Setting Data With Accessors
You can use accessors to modify parts of a Pandas DataFrame by passing a Python sequence, NumPy array, or single value:
>>>
>>> df . loc [:, 'py-score' ] x 88.0 xi 79.0 12 81.0 thirteen 80.0 fourteen 68.0 15 61.0 16 84.0 Proper name: py-score, dtype: float64 >>> df . loc [: 13 , 'py-score' ] = [ twoscore , 50 , 60 , 70 ] >>> df . loc [ xiv :, 'py-score' ] = 0 >>> df [ 'py-score' ] 10 40.0 11 fifty.0 12 threescore.0 13 70.0 xiv 0.0 15 0.0 16 0.0 Name: py-score, dtype: float64 The statement df.loc[:xiii, 'py-score'] = [40, fifty, 60, 70] modifies the first four items (rows 10 through 13) in the column py-score using the values from your supplied listing. Using df.loc[14:, 'py-score'] = 0 sets the remaining values in this column to 0.
The following instance shows that you can use negative indices with .iloc[] to admission or modify information:
>>>
>>> df . iloc [:, - 1 ] = np . array ([ 88.0 , 79.0 , 81.0 , 80.0 , 68.0 , 61.0 , 84.0 ]) >>> df [ 'py-score' ] 10 88.0 11 79.0 12 81.0 13 eighty.0 14 68.0 15 61.0 16 84.0 Name: py-score, dtype: float64 In this example, you've accessed and modified the last column ('py-score'), which corresponds to the integer cavalcade index -1. This behavior is consistent with Python sequences and NumPy arrays.
Inserting and Deleting Data
Pandas provides several user-friendly techniques for inserting and deleting rows or columns. You tin cull among them based on your situation and needs.
Inserting and Deleting Rows
Imagine you want to add together a new person to your list of job candidates. You can start past creating a new Series object that represents this new candidate:
>>>
>>> john = pd . Series ( data = [ 'John' , 'Boston' , 34 , 79 ], ... alphabetize = df . columns , name = 17 ) >>> john name John city Boston historic period 34 py-score 79 Name: 17, dtype: object >>> john . proper name 17 The new object has labels that correspond to the column labels from df. That's why you lot demand index=df.columns.
You tin add john as a new row to the end of df with .append():
>>>
>>> df = df . append ( john ) >>> df proper noun urban center age py-score 10 Xavier Mexico City 41 88.0 11 Ann Toronto 28 79.0 12 Jana Prague 33 81.0 xiii Yi Shanghai 34 lxxx.0 14 Robin Manchester 38 68.0 fifteen Amal Cairo 31 61.0 xvi Nori Osaka 37 84.0 17 John Boston 34 79.0 Here, .append() returns the Pandas DataFrame with the new row appended. Notice how Pandas uses the attribute john.name, which is the value 17, to specify the characterization for the new row.
You've appended a new row with a single call to .append(), and you lot can delete it with a single call to .drop():
>>>
>>> df = df . drop ( labels = [ 17 ]) >>> df name city age py-score 10 Xavier Mexico Urban center 41 88.0 11 Ann Toronto 28 79.0 12 Jana Prague 33 81.0 xiii Yi Shanghai 34 80.0 fourteen Robin Manchester 38 68.0 xv Amal Cairo 31 61.0 sixteen Nori Osaka 37 84.0 Hither, .drop() removes the rows specified with the parameter labels. By default, it returns the Pandas DataFrame with the specified rows removed. If you lot pass inplace=True, so the original DataFrame volition be modified and you'll get None as the return value.
Inserting and Deleting Columns
The nearly straightforward way to insert a column in a Pandas DataFrame is to follow the aforementioned procedure that you utilize when yous add together an item to a dictionary. Here's how you can suspend a cavalcade containing your candidates' scores on a JavaScript test:
>>>
>>> df [ 'js-score' ] = np . array ([ 71.0 , 95.0 , 88.0 , 79.0 , 91.0 , 91.0 , 80.0 ]) >>> df name city historic period py-score js-score ten Xavier Mexico City 41 88.0 71.0 11 Ann Toronto 28 79.0 95.0 12 Jana Prague 33 81.0 88.0 13 Yi Shanghai 34 80.0 79.0 xiv Robin Manchester 38 68.0 91.0 15 Amal Cairo 31 61.0 91.0 16 Nori Osaka 37 84.0 eighty.0 Now the original DataFrame has one more column, js-score, at its end.
You don't have to provide a total sequence of values. You can add a new column with a single value:
>>>
>>> df [ 'total-score' ] = 0.0 >>> df name city age py-score js-score full-score 10 Xavier United mexican states City 41 88.0 71.0 0.0 11 Ann Toronto 28 79.0 95.0 0.0 12 Jana Prague 33 81.0 88.0 0.0 13 Yi Shanghai 34 fourscore.0 79.0 0.0 xiv Robin Manchester 38 68.0 91.0 0.0 xv Amal Cairo 31 61.0 91.0 0.0 xvi Nori Osaka 37 84.0 80.0 0.0 The DataFrame df now has an additional cavalcade filled with zeros.
If you've used dictionaries in the past, then this way of inserting columns might be familiar to you. However, it doesn't allow y'all to specify the location of the new cavalcade. If the location of the new column is important, then you tin use .insert() instead:
>>>
>>> df . insert ( loc = four , column = 'django-score' , ... value = np . array ([ 86.0 , 81.0 , 78.0 , 88.0 , 74.0 , 70.0 , 81.0 ])) >>> df name city age py-score django-score js-score total-score 10 Xavier Mexico City 41 88.0 86.0 71.0 0.0 eleven Ann Toronto 28 79.0 81.0 95.0 0.0 12 Jana Prague 33 81.0 78.0 88.0 0.0 13 Yi Shanghai 34 80.0 88.0 79.0 0.0 14 Robin Manchester 38 68.0 74.0 91.0 0.0 15 Amal Cairo 31 61.0 70.0 91.0 0.0 xvi Nori Osaka 37 84.0 81.0 80.0 0.0 You've just inserted another column with the score of the Django examination. The parameter loc determines the location, or the zero-based alphabetize, of the new column in the Pandas DataFrame. column sets the label of the new column, and value specifies the data values to insert.
You can delete one or more columns from a Pandas DataFrame just as you would with a regular Python dictionary, past using the del statement:
>>>
>>> del df [ 'total-score' ] >>> df name city age py-score django-score js-score 10 Xavier Mexico City 41 88.0 86.0 71.0 xi Ann Toronto 28 79.0 81.0 95.0 12 Jana Prague 33 81.0 78.0 88.0 13 Yi Shanghai 34 80.0 88.0 79.0 14 Robin Manchester 38 68.0 74.0 91.0 15 Amal Cairo 31 61.0 lxx.0 91.0 16 Nori Osaka 37 84.0 81.0 80.0 Now you take df without the column total-score. Another similarity to dictionaries is the power to use .pop(), which removes the specified cavalcade and returns it. That means you could do something like df.popular('total-score') instead of using del.
You can too remove one or more columns with .drop() as you did previously with the rows. Again, you demand to specify the labels of the desired columns with labels. In addition, when you lot want to remove columns, you need to provide the argument axis=1:
>>>
>>> df = df . drib ( labels = 'age' , axis = ane ) >>> df proper noun metropolis py-score django-score js-score ten Xavier Mexico City 88.0 86.0 71.0 11 Ann Toronto 79.0 81.0 95.0 12 Jana Prague 81.0 78.0 88.0 13 Yi Shanghai 80.0 88.0 79.0 14 Robin Manchester 68.0 74.0 91.0 xv Amal Cairo 61.0 70.0 91.0 16 Nori Osaka 84.0 81.0 80.0 You lot've removed the cavalcade age from your DataFrame.
By default, .drib() returns the DataFrame without the specified columns unless yous pass inplace=True.
Applying Arithmetic Operations
Yous can utilise basic arithmetic operations such equally addition, subtraction, multiplication, and partitioning to Pandas Series and DataFrame objects the same manner you would with NumPy arrays:
>>>
>>> df [ 'py-score' ] + df [ 'js-score' ] 10 159.0 11 174.0 12 169.0 thirteen 159.0 14 159.0 15 152.0 16 164.0 dtype: float64 >>> df [ 'py-score' ] / 100 x 0.88 11 0.79 12 0.81 13 0.80 14 0.68 15 0.61 16 0.84 Name: py-score, dtype: float64 You lot tin use this technique to insert a new column to a Pandas DataFrame. For instance, attempt calculating a total score as a linear combination of your candidates' Python, Django, and JavaScript scores:
>>>
>>> df [ 'full' ] =\ ... 0.four * df [ 'py-score' ] + 0.iii * df [ 'django-score' ] + 0.3 * df [ 'js-score' ] >>> df name city py-score django-score js-score total 10 Xavier Mexico City 88.0 86.0 71.0 82.iii 11 Ann Toronto 79.0 81.0 95.0 84.4 12 Jana Prague 81.0 78.0 88.0 82.2 13 Yi Shanghai 80.0 88.0 79.0 82.1 fourteen Robin Manchester 68.0 74.0 91.0 76.7 15 Amal Cairo 61.0 lxx.0 91.0 72.7 sixteen Nori Osaka 84.0 81.0 80.0 81.9 At present your DataFrame has a column with a total score calculated from your candidates' individual test scores. Even amend, you lot accomplished that with just a single statement!
Applying NumPy and SciPy Functions
Most NumPy and SciPy routines can be applied to Pandas Series or DataFrame objects as arguments instead of as NumPy arrays. To illustrate this, you can calculate candidates' total examination scores using the NumPy routine numpy.average().
Instead of passing a NumPy assortment to numpy.boilerplate(), you'll laissez passer a role of your Pandas DataFrame:
>>>
>>> import numpy equally np >>> score = df . iloc [:, 2 : five ] >>> score py-score django-score js-score 10 88.0 86.0 71.0 11 79.0 81.0 95.0 12 81.0 78.0 88.0 13 eighty.0 88.0 79.0 xiv 68.0 74.0 91.0 15 61.0 seventy.0 91.0 16 84.0 81.0 eighty.0 >>> np . average ( score , axis = 1 , ... weights = [ 0.4 , 0.3 , 0.iii ]) array([82.3, 84.4, 82.2, 82.i, 76.7, 72.vii, 81.nine]) The variable score now refers to the DataFrame with the Python, Django, and JavaScript scores. Yous tin can use score equally an statement of numpy.average() and get the linear combination of columns with the specified weights.
But that'south not all! You can use the NumPy array returned past average() as a new cavalcade of df. First, delete the existing column total from df, and so suspend the new one using average():
>>>
>>> del df [ 'total' ] >>> df proper name metropolis py-score django-score js-score 10 Xavier Mexico Metropolis 88.0 86.0 71.0 11 Ann Toronto 79.0 81.0 95.0 12 Jana Prague 81.0 78.0 88.0 13 Yi Shanghai 80.0 88.0 79.0 xiv Robin Manchester 68.0 74.0 91.0 fifteen Amal Cairo 61.0 70.0 91.0 sixteen Nori Osaka 84.0 81.0 lxxx.0 >>> df [ 'total' ] = np . average ( df . iloc [:, two : 5 ], axis = 1 , ... weights = [ 0.iv , 0.three , 0.three ]) >>> df proper noun city py-score django-score js-score total x Xavier Mexico Urban center 88.0 86.0 71.0 82.3 11 Ann Toronto 79.0 81.0 95.0 84.4 12 Jana Prague 81.0 78.0 88.0 82.2 thirteen Yi Shanghai 80.0 88.0 79.0 82.1 14 Robin Manchester 68.0 74.0 91.0 76.vii 15 Amal Cairo 61.0 70.0 91.0 72.7 sixteen Nori Osaka 84.0 81.0 lxxx.0 81.9 The result is the aforementioned as in the previous example, simply hither you used the existing NumPy role instead of writing your ain lawmaking.
Sorting a Pandas DataFrame
Y'all can sort a Pandas DataFrame with .sort_values():
>>>
>>> df . sort_values ( past = 'js-score' , ascending = Simulated ) proper name metropolis py-score django-score js-score total eleven Ann Toronto 79.0 81.0 95.0 84.4 14 Robin Manchester 68.0 74.0 91.0 76.7 fifteen Amal Cairo 61.0 lxx.0 91.0 72.seven 12 Jana Prague 81.0 78.0 88.0 82.two 16 Nori Osaka 84.0 81.0 80.0 81.nine 13 Yi Shanghai 80.0 88.0 79.0 82.1 x Xavier Mexico City 88.0 86.0 71.0 82.three This example sorts your DataFrame by the values in the cavalcade js-score. The parameter by sets the label of the row or column to sort by. ascending specifies whether you want to sort in ascending (True) or descending (Simulated) guild, the latter existence the default setting. You can pass centrality to choose if you want to sort rows (axis=0) or columns (axis=i).
If y'all want to sort past multiple columns, so only pass lists as arguments for past and ascending:
>>>
>>> df . sort_values ( past = [ 'total' , 'py-score' ], ascending = [ False , False ]) proper name urban center py-score django-score js-score total 11 Ann Toronto 79.0 81.0 95.0 84.four 10 Xavier Mexico City 88.0 86.0 71.0 82.3 12 Jana Prague 81.0 78.0 88.0 82.2 13 Yi Shanghai 80.0 88.0 79.0 82.1 16 Nori Osaka 84.0 81.0 80.0 81.9 14 Robin Manchester 68.0 74.0 91.0 76.7 fifteen Amal Cairo 61.0 70.0 91.0 72.vii In this instance, the DataFrame is sorted by the cavalcade full, simply if two values are the same, then their order is determined past the values from the cavalcade py-score.
The optional parameter inplace can besides be used with .sort_values(). Information technology'due south set to False by default, ensuring .sort_values() returns a new Pandas DataFrame. When y'all set inplace=True, the existing DataFrame will be modified and .sort_values() will return None.
If you lot've e'er tried to sort values in Excel, so you lot might detect the Pandas arroyo much more efficient and convenient. When y'all have large amounts of data, Pandas can significantly outperform Excel.
For more than information on sorting in Pandas, check out Pandas Sort: Your Guide to Sorting Information in Python.
Filtering Information
Information filtering is another powerful characteristic of Pandas. It works similarly to indexing with Boolean arrays in NumPy.
If you apply some logical operation on a Series object, then y'all'll get another Series with the Boolean values True and False:
>>>
>>> filter_ = df [ 'django-score' ] >= eighty >>> filter_ 10 True 11 True 12 False 13 Truthful 14 False 15 Faux 16 Truthful Name: django-score, dtype: bool In this case, df['django-score'] >= lxxx returns True for those rows in which the Django score is greater than or equal to eighty. It returns False for the rows with a Django score less than 80.
You lot now have the Series filter_ filled with Boolean data. The expression df[filter_] returns a Pandas DataFrame with the rows from df that correspond to True in filter_:
>>>
>>> df [ filter_ ] proper name city py-score django-score js-score total ten Xavier United mexican states City 88.0 86.0 71.0 82.3 11 Ann Toronto 79.0 81.0 95.0 84.4 xiii Yi Shanghai eighty.0 88.0 79.0 82.i 16 Nori Osaka 84.0 81.0 80.0 81.9 As you can see, filter_[10], filter_[11], filter_[xiii], and filter_[16] are True, so df[filter_] contains the rows with these labels. On the other hand, filter_[12], filter_[14], and filter_[15] are Faux, so the respective rows don't appear in df[filter_].
Y'all can create very powerful and sophisticated expressions by combining logical operations with the following operators:
-
NOT(~) -
AND(&) -
OR(|) -
XOR(^)
For example, you can get a DataFrame with the candidates whose py-score and js-score are greater than or equal to 80:
>>>
>>> df [( df [ 'py-score' ] >= 80 ) & ( df [ 'js-score' ] >= 80 )] name city py-score django-score js-score full 12 Jana Prague 81.0 78.0 88.0 82.two 16 Nori Osaka 84.0 81.0 lxxx.0 81.9 The expression (df['py-score'] >= eighty) & (df['js-score'] >= 80) returns a Series with True in the rows for which both py-score and js-score are greater than or equal to lxxx and Imitation in the others. In this case, only the rows with the labels 12 and 16 satisfy both conditions.
You tin can also use NumPy logical routines instead of operators.
For some operations that require data filtering, it's more than convenient to apply .where(). It replaces the values in the positions where the provided condition isn't satisfied:
>>>
>>> df [ 'django-score' ] . where ( cond = df [ 'django-score' ] >= 80 , other = 0.0 ) x 86.0 eleven 81.0 12 0.0 xiii 88.0 14 0.0 fifteen 0.0 xvi 81.0 Proper noun: django-score, dtype: float64 In this example, the condition is df['django-score'] >= 80. The values of the DataFrame or Series that calls .where() will remain the same where the condition is True and will be replaced with the value of other (in this case 0.0) where the condition is False.
Determining Data Statistics
Pandas provides many statistical methods for DataFrames. You tin get basic statistics for the numerical columns of a Pandas DataFrame with .describe():
>>>
>>> df . describe () py-score django-score js-score full count seven.000000 seven.000000 7.000000 7.000000 mean 77.285714 79.714286 85.000000 80.328571 std nine.446592 six.343350 8.544004 4.101510 min 61.000000 seventy.000000 71.000000 72.700000 25% 73.500000 76.000000 79.500000 79.300000 fifty% eighty.000000 81.000000 88.000000 82.100000 75% 82.500000 83.500000 91.000000 82.250000 max 88.000000 88.000000 95.000000 84.400000 Here, .describe() returns a new DataFrame with the number of rows indicated by count, as well every bit the hateful, standard deviation, minimum, maximum, and quartiles of the columns.
If you want to become detail statistics for some or all of your columns, and so y'all can call methods such as .hateful() or .std():
>>>
>>> df . hateful () py-score 77.285714 django-score 79.714286 js-score 85.000000 full 80.328571 dtype: float64 >>> df [ 'py-score' ] . mean () 77.28571428571429 >>> df . std () py-score nine.446592 django-score 6.343350 js-score 8.544004 total 4.101510 dtype: float64 >>> df [ 'py-score' ] . std () 9.446591726019244 When practical to a Pandas DataFrame, these methods return Series with the results for each column. When applied to a Series object, or a single column of a DataFrame, the methods return scalars.
To learn more about statistical calculations with Pandas, check out Descriptive Statistics With Python and NumPy, SciPy, and Pandas: Correlation With Python.
Handling Missing Data
Missing data is very common in data science and motorcar learning. Simply never fearfulness! Pandas has very powerful features for working with missing data. In fact, its documentation has an entire section dedicated to working with missing data.
Pandas usually represents missing data with NaN (not a number) values. In Python, you can go NaN with float('nan'), math.nan, or numpy.nan. Starting with Pandas 1.0, newer types like BooleanDtype, Int8Dtype, Int16Dtype, Int32Dtype, and Int64Dtype utilize pandas.NA as a missing value.
Here'southward an example of a Pandas DataFrame with a missing value:
>>>
>>> df_ = pd . DataFrame ({ '10' : [ 1 , two , np . nan , iv ]}) >>> df_ ten 0 one.0 one 2.0 2 NaN 3 four.0 The variable df_ refers to the DataFrame with one column, x, and four values. The third value is nan and is considered missing by default.
Computing With Missing Data
Many Pandas methods omit nan values when performing calculations unless they are explicitly instructed not to:
>>>
>>> df_ . mean () 10 2.333333 dtype: float64 >>> df_ . hateful ( skipna = Faux ) 10 NaN dtype: float64 In the starting time example, df_.mean() calculates the mean without taking NaN (the 3rd value) into business relationship. It but takes ane.0, 2.0, and 4.0 and returns their average, which is 2.33.
However, if you instruct .mean() non to skip nan values with skipna=False, then it will consider them and return nan if there's whatsoever missing value amongst the data.
Filling Missing Data
Pandas has several options for filling, or replacing, missing values with other values. One of the well-nigh convenient methods is .fillna(). You can employ it to replace missing values with:
- Specified values
- The values above the missing value
- The values below the missing value
Here's how you lot can apply the options mentioned higher up:
>>>
>>> df_ . fillna ( value = 0 ) x 0 one.0 i 2.0 ii 0.0 three iv.0 >>> df_ . fillna ( method = 'ffill' ) x 0 1.0 1 2.0 2 2.0 iii 4.0 >>> df_ . fillna ( method = 'bfill' ) 10 0 1.0 1 2.0 2 4.0 3 4.0 In the first example, .fillna(value=0) replaces the missing value with 0.0, which you specified with value. In the 2nd case, .fillna(method='ffill') replaces the missing value with the value above it, which is ii.0. In the third example, .fillna(method='bfill') uses the value beneath the missing value, which is four.0.
Some other pop option is to apply interpolation and supplant missing values with interpolated values. You can do this with .interpolate():
>>>
>>> df_ . interpolate () x 0 ane.0 one two.0 two 3.0 three 4.0 Every bit you lot can come across, .interpolate() replaces the missing value with an interpolated value.
You tin can also utilize the optional parameter inplace with .fillna(). Doing so volition:
- Create and render a new DataFrame when
inplace=False - Change the existing DataFrame and return
Nonewheninplace=True
The default setting for inplace is Faux. However, inplace=True can be very useful when you're working with large amounts of data and want to prevent unnecessary and inefficient copying.
Deleting Rows and Columns With Missing Information
In certain situations, yous might want to delete rows or even columns that have missing values. You lot tin do this with .dropna():
>>>
>>> df_ . dropna () x 0 1.0 ane 2.0 3 four.0 In this case, .dropna() simply deletes the row with nan, including its characterization. It also has the optional parameter inplace, which behaves the same as it does with .fillna() and .interpolate().
Iterating Over a Pandas DataFrame
Every bit you learned earlier, a DataFrame'south row and column labels tin can exist retrieved as sequences with .index and .columns. You can utilize this feature to iterate over labels and get or fix information values. All the same, Pandas provides several more than user-friendly methods for iteration:
-
.items()to iterate over columns -
.iteritems()to iterate over columns -
.iterrows()to iterate over rows -
.itertuples()to iterate over rows and go named tuples
With .items() and .iteritems(), you iterate over the columns of a Pandas DataFrame. Each iteration yields a tuple with the name of the column and the cavalcade information as a Series object:
>>>
>>> for col_label , col in df . iteritems (): ... print ( col_label , col , sep = ' \n ' , stop = ' \n\northward ' ) ... name ten Xavier eleven Ann 12 Jana thirteen Yi 14 Robin 15 Amal 16 Nori Name: name, dtype: object urban center 10 Mexico Urban center eleven Toronto 12 Prague xiii Shanghai 14 Manchester xv Cairo xvi Osaka Name: urban center, dtype: object py-score 10 88.0 xi 79.0 12 81.0 13 80.0 14 68.0 fifteen 61.0 16 84.0 Name: py-score, dtype: float64 django-score 10 86.0 11 81.0 12 78.0 13 88.0 xiv 74.0 15 seventy.0 16 81.0 Proper noun: django-score, dtype: float64 js-score ten 71.0 11 95.0 12 88.0 thirteen 79.0 fourteen 91.0 15 91.0 sixteen 80.0 Proper noun: js-score, dtype: float64 total ten 82.3 11 84.4 12 82.2 13 82.one fourteen 76.7 15 72.vii sixteen 81.ix Proper name: total, dtype: float64 That's how you use .items() and .iteritems().
With .iterrows(), y'all iterate over the rows of a Pandas DataFrame. Each iteration yields a tuple with the name of the row and the row data equally a Series object:
>>>
>>> for row_label , row in df . iterrows (): ... impress ( row_label , row , sep = ' \n ' , finish = ' \due north\due north ' ) ... ten name Xavier city United mexican states City py-score 88 django-score 86 js-score 71 total 82.3 Proper name: 10, dtype: object 11 name Ann city Toronto py-score 79 django-score 81 js-score 95 total 84.four Proper noun: 11, dtype: object 12 name Jana metropolis Prague py-score 81 django-score 78 js-score 88 total 82.2 Proper name: 12, dtype: object thirteen name Yi metropolis Shanghai py-score lxxx django-score 88 js-score 79 full 82.1 Proper noun: 13, dtype: object fourteen name Robin city Manchester py-score 68 django-score 74 js-score 91 full 76.7 Proper noun: 14, dtype: object 15 name Amal city Cairo py-score 61 django-score 70 js-score 91 total 72.7 Proper name: fifteen, dtype: object xvi proper name Nori city Osaka py-score 84 django-score 81 js-score eighty full 81.9 Proper noun: sixteen, dtype: object That's how you apply .iterrows().
Similarly, .itertuples() iterates over the rows and in each iteration yields a named tuple with (optionally) the index and data:
>>>
>>> for row in df . loc [:, [ 'proper name' , 'city' , 'total' ]] . itertuples (): ... print ( row ) ... Pandas(Index=10, name='Xavier', metropolis='Mexico City', total=82.3) Pandas(Alphabetize=eleven, name='Ann', city='Toronto', total=84.4) Pandas(Alphabetize=12, name='Jana', city='Prague', total=82.19999999999999) Pandas(Index=13, name='Yi', metropolis='Shanghai', total=82.i) Pandas(Index=14, name='Robin', city='Manchester', total=76.7) Pandas(Index=fifteen, name='Amal', city='Cairo', full=72.7) Pandas(Index=xvi, proper name='Nori', city='Osaka', full=81.9) Yous tin specify the name of the named tuple with the parameter name, which is prepare to 'Pandas' past default. Yous can also specify whether to include row labels with alphabetize, which is set to Truthful by default.
Working With Time Series
Pandas excels at handling time serial. Although this functionality is partly based on NumPy datetimes and timedeltas, Pandas provides much more flexibility.
Creating DataFrames With Time-Serial Labels
In this section, you'll create a Pandas DataFrame using the hourly temperature data from a single day.
Y'all can offset by creating a list (or tuple, NumPy assortment, or other data type) with the data values, which will be hourly temperatures given in degrees Celsius:
>>>
>>> temp_c = [ 8.0 , 7.1 , half-dozen.8 , 6.iv , vi.0 , v.4 , iv.8 , five.0 , ... 9.1 , 12.8 , fifteen.3 , nineteen.ane , 21.2 , 22.1 , 22.4 , 23.1 , ... 21.0 , 17.9 , 15.five , 14.4 , 11.ix , xi.0 , ten.2 , nine.1 ] Now you lot have the variable temp_c, which refers to the list of temperature values.
The side by side step is to create a sequence of dates and times. Pandas provides a very convenient function, date_range(), for this purpose:
>>>
>>> dt = pd . date_range ( kickoff = '2019-10-27 00:00:00.0' , periods = 24 , ... freq = 'H' ) >>> dt DatetimeIndex(['2019-10-27 00:00:00', '2019-10-27 01:00:00', '2019-10-27 02:00:00', '2019-ten-27 03:00:00', '2019-10-27 04:00:00', '2019-10-27 05:00:00', '2019-10-27 06:00:00', '2019-10-27 07:00:00', '2019-10-27 08:00:00', '2019-x-27 09:00:00', '2019-10-27 10:00:00', '2019-10-27 11:00:00', '2019-x-27 12:00:00', '2019-10-27 thirteen:00:00', '2019-10-27 xiv:00:00', '2019-x-27 15:00:00', '2019-x-27 xvi:00:00', '2019-10-27 17:00:00', '2019-10-27 18:00:00', '2019-10-27 19:00:00', '2019-ten-27 20:00:00', '2019-10-27 21:00:00', '2019-10-27 22:00:00', '2019-10-27 23:00:00'], dtype='datetime64[ns]', freq='H') date_range() accepts the arguments that you use to specify the start or end of the range, number of periods, frequency, fourth dimension zone, and more than.
Now that you have the temperature values and the corresponding dates and times, yous tin create the DataFrame. In many cases, it'due south convenient to use engagement-time values every bit the row labels:
>>>
>>> temp = pd . DataFrame ( data = { 'temp_c' : temp_c }, alphabetize = dt ) >>> temp temp_c 2019-10-27 00:00:00 8.0 2019-10-27 01:00:00 seven.one 2019-10-27 02:00:00 6.8 2019-10-27 03:00:00 6.4 2019-10-27 04:00:00 6.0 2019-ten-27 05:00:00 five.4 2019-10-27 06:00:00 4.8 2019-x-27 07:00:00 five.0 2019-10-27 08:00:00 ix.i 2019-10-27 09:00:00 12.8 2019-x-27 ten:00:00 15.three 2019-x-27 xi:00:00 19.i 2019-x-27 12:00:00 21.ii 2019-ten-27 13:00:00 22.1 2019-10-27 14:00:00 22.4 2019-10-27 15:00:00 23.one 2019-ten-27 16:00:00 21.0 2019-x-27 17:00:00 17.ix 2019-10-27 eighteen:00:00 15.5 2019-10-27 nineteen:00:00 14.4 2019-10-27 xx:00:00 xi.nine 2019-10-27 21:00:00 11.0 2019-10-27 22:00:00 ten.2 2019-10-27 23:00:00 9.one That'due south it! You've created a DataFrame with fourth dimension-series data and engagement-time row indices.
Indexing and Slicing
Once you have a Pandas DataFrame with time-series information, y'all can conveniently utilize slicing to get merely a part of the information:
>>>
>>> temp [ '2019-10-27 05' : '2019-ten-27 14' ] temp_c 2019-10-27 05:00:00 five.four 2019-10-27 06:00:00 four.viii 2019-10-27 07:00:00 v.0 2019-10-27 08:00:00 9.1 2019-10-27 09:00:00 12.eight 2019-10-27 ten:00:00 15.iii 2019-10-27 11:00:00 19.one 2019-x-27 12:00:00 21.2 2019-10-27 thirteen:00:00 22.1 2019-10-27 xiv:00:00 22.four This example shows how to excerpt the temperatures between 05:00 and 14:00 (5 a.1000. and 2 p.m.). Although you've provided strings, Pandas knows that your row labels are appointment-time values and interprets the strings as dates and times.
Resampling and Rolling
You lot've just seen how to combine engagement-time row labels and employ slicing to get the data you need from the time-series data. This is just the beginning. Information technology gets better!
If you want to divide a 24-hour interval into four half-dozen-hour intervals and get the hateful temperature for each interval, and so you're simply ane argument away from doing so. Pandas provides the method .resample(), which you can combine with other methods such as .mean():
>>>
>>> temp . resample ( rule = '6h' ) . mean () temp_c 2019-10-27 00:00:00 half dozen.616667 2019-10-27 06:00:00 11.016667 2019-ten-27 12:00:00 21.283333 2019-10-27 18:00:00 12.016667 Y'all at present have a new Pandas DataFrame with four rows. Each row corresponds to a unmarried half-dozen-hour interval. For example, the value 6.616667 is the mean of the first six temperatures from the DataFrame temp, whereas 12.016667 is the mean of the concluding six temperatures.
Instead of .hateful(), you can apply .min() or .max() to get the minimum and maximum temperatures for each interval. You lot can also use .sum() to get the sums of data values, although this information probably isn't useful when you're working with temperatures.
You might also need to do some rolling-window analysis. This involves computing a statistic for a specified number of adjacent rows, which make up your window of information. You can "coil" the window by selecting a different prepare of adjacent rows to perform your calculations on.
Your kickoff window starts with the first row in your DataFrame and includes as many adjacent rows every bit y'all specify. You then move your window downward 1 row, dropping the first row and adding the row that comes immediately afterwards the terminal row, and calculate the aforementioned statistic again. You repeat this process until you attain the concluding row of the DataFrame.
Pandas provides the method .rolling() for this purpose:
>>>
>>> temp . rolling ( window = three ) . mean () temp_c 2019-10-27 00:00:00 NaN 2019-x-27 01:00:00 NaN 2019-ten-27 02:00:00 7.300000 2019-10-27 03:00:00 6.766667 2019-10-27 04:00:00 6.400000 2019-10-27 05:00:00 5.933333 2019-x-27 06:00:00 5.400000 2019-ten-27 07:00:00 five.066667 2019-10-27 08:00:00 six.300000 2019-10-27 09:00:00 viii.966667 2019-10-27 10:00:00 12.400000 2019-10-27 11:00:00 xv.733333 2019-10-27 12:00:00 xviii.533333 2019-10-27 13:00:00 20.800000 2019-10-27 14:00:00 21.900000 2019-10-27 15:00:00 22.533333 2019-10-27 xvi:00:00 22.166667 2019-10-27 17:00:00 twenty.666667 2019-10-27 eighteen:00:00 18.133333 2019-10-27 xix:00:00 15.933333 2019-10-27 xx:00:00 13.933333 2019-10-27 21:00:00 12.433333 2019-10-27 22:00:00 11.033333 2019-ten-27 23:00:00 ten.100000 Now you lot have a DataFrame with mean temperatures calculated for several three-hour windows. The parameter window specifies the size of the moving time window.
In the example above, the tertiary value (seven.3) is the mean temperature for the first three hours (00:00:00, 01:00:00, and 02:00:00). The fourth value is the mean temperature for the hours 02:00:00, 03:00:00, and 04:00:00. The final value is the mean temperature for the concluding three hours, 21:00:00, 22:00:00, and 23:00:00. The first two values are missing because there isn't plenty data to calculate them.
Plotting With Pandas DataFrames
Pandas allows you to visualize data or create plots based on DataFrames. It uses Matplotlib in the background, so exploiting Pandas' plotting capabilities is very similar to working with Matplotlib.
If you desire to brandish the plots, then you lot first demand to import matplotlib.pyplot:
>>>
>>> import matplotlib.pyplot as plt Now y'all can use pandas.DataFrame.plot() to create the plot and plt.show() to display information technology:
>>>
>>> temp . plot () <matplotlib.axes._subplots.AxesSubplot object at 0x7f070cd9d950> >>> plt . testify () Now .plot() returns a plot object that looks like this:
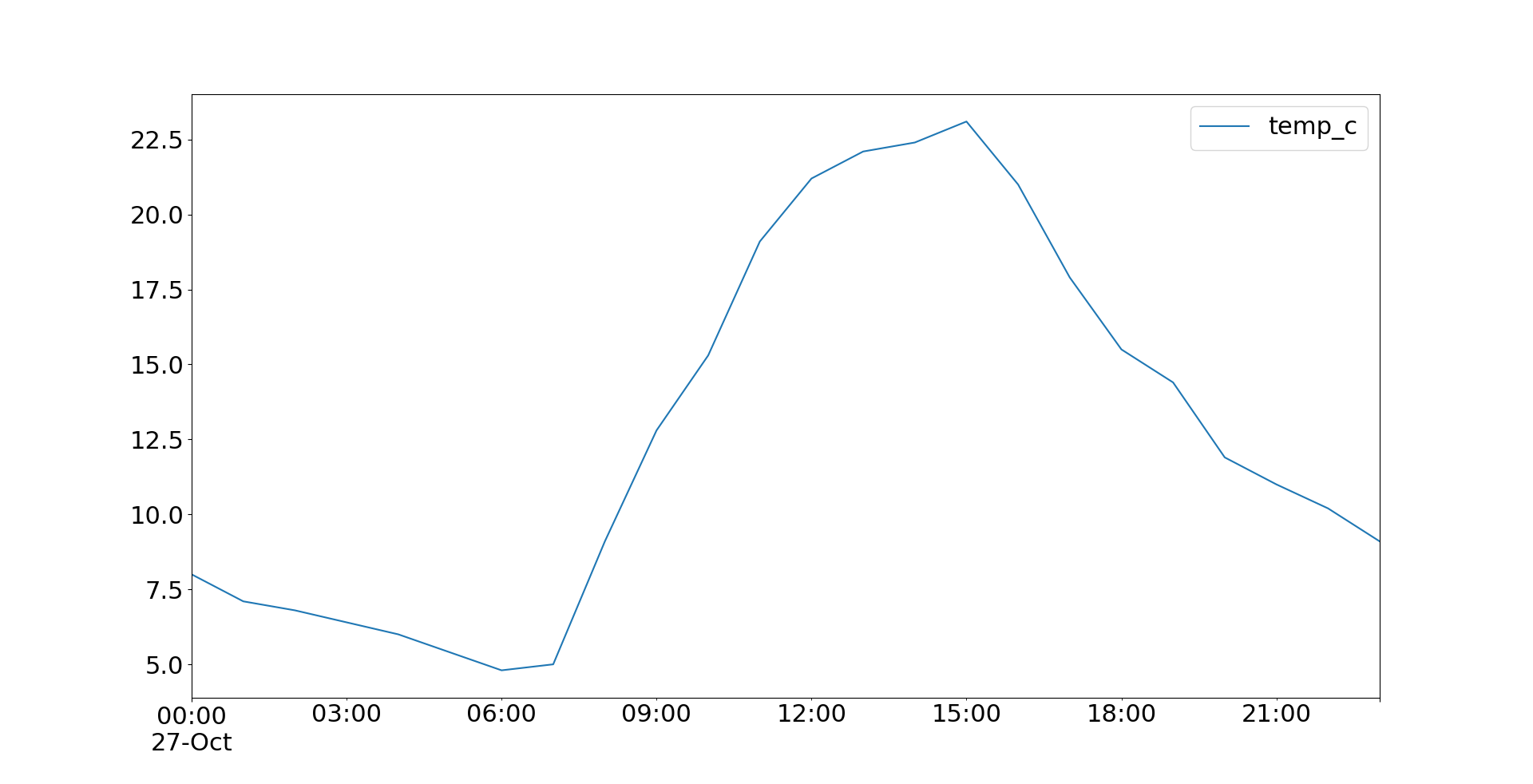
You can also utilise .plot.line() and become the same result. Both .plot() and .plot.line() have many optional parameters that you can use to specify the look of your plot. Some of them are passed directly to the underlying Matplotlib methods.
You can save your figure past chaining the methods .get_figure() and .savefig():
>>>
>>> temp . plot () . get_figure () . savefig ( 'temperatures.png' ) This argument creates the plot and saves it as a file called 'temperatures.png' in your working directory.
Y'all can go other types of plots with a Pandas DataFrame. For example, you can visualize your job candidate data from earlier every bit a histogram with .plot.hist():
>>>
>>> df . loc [:, [ 'py-score' , 'total' ]] . plot . hist ( bins = v , alpha = 0.4 ) <matplotlib.axes._subplots.AxesSubplot object at 0x7f070c69edd0> >>> plt . show () In this example, you extract the Python examination score and total score data and visualize it with a histogram. The resulting plot looks like this:
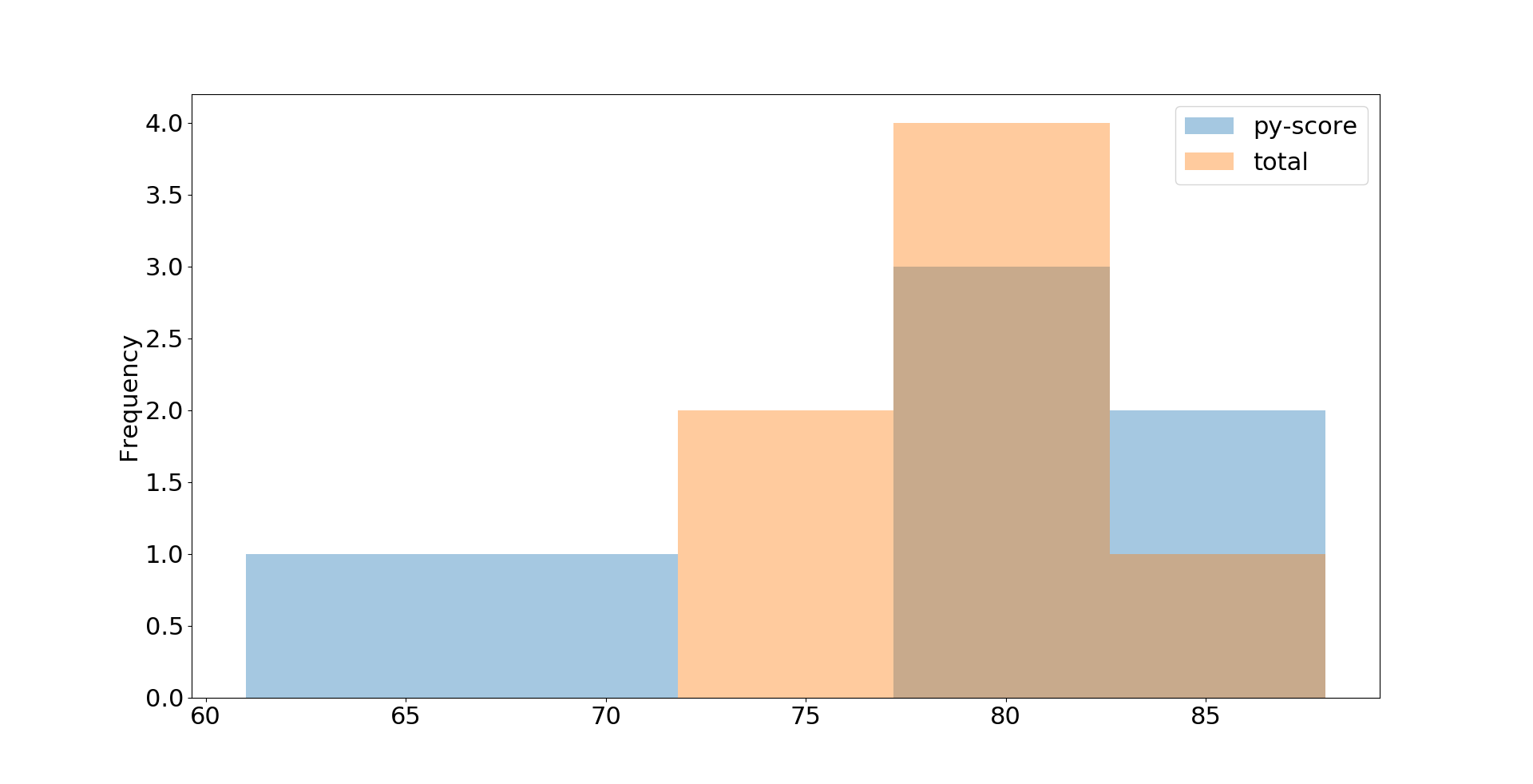
This is just the bones look. You can suit details with optional parameters including .plot.hist(), Matplotlib'south plt.rcParams, and many others. You can find detailed explanations in the Anatomy of Matplotlib.
Farther Reading
Pandas DataFrames are very comprehensive objects that support many operations not mentioned in this tutorial. Some of these include:
- Hierarchical (multi-level) indexing
- Grouping
- Merging, joining, and concatenating
- Working with categorical data
The official Pandas tutorial summarizes some of the bachelor options nicely. If you want to learn more than almost Pandas and DataFrames, and then you tin check out these tutorials:
- Pythonic Information Cleaning With Pandas and NumPy
- Pandas DataFrames 101
- Introduction to Pandas and Vincent
- Python Pandas: Tricks & Features Yous May Not Know
- Idiomatic Pandas: Tricks & Features Y'all May Not Know
- Reading CSVs With Pandas
- Writing CSVs With Pandas
- Reading and Writing CSV Files in Python
- Reading and Writing CSV Files
- Using Pandas to Read Big Excel Files in Python
- Fast, Flexible, Easy and Intuitive: How to Speed Up Your Pandas Projects
You've learned that Pandas DataFrames handle two-dimensional data. If you need to work with labeled data in more than than two dimensions, you tin check out xarray, some other powerful Python library for information science with very similar features to Pandas.
If y'all piece of work with large data and want a DataFrame-similar experience, then you might give Dask a risk and use its DataFrame API. A Dask DataFrame contains many Pandas DataFrames and performs computations in a lazy manner.
Conclusion
You now know what a Pandas DataFrame is, what some of its features are, and how you can use it to work with data efficiently. Pandas DataFrames are powerful, convenient data structures that you tin use to proceeds deeper insight into your datasets!
In this tutorial, you lot've learned:
- What a Pandas DataFrame is and how to create one
- How to admission, modify, add, sort, filter, and delete data
- How to use NumPy routines with DataFrames
- How to handle missing values
- How to piece of work with fourth dimension-series data
- How to visualize information independent in DataFrames
Y'all've learned enough to cover the fundamentals of DataFrames. If y'all want to dig deeper into working with data in Python, and then check out the entire range of Pandas tutorials.
If y'all have questions or comments, and so delight put them in the comment section beneath.
Watch Now This tutorial has a related video course created by the Real Python team. Watch information technology together with the written tutorial to deepen your understanding: The Pandas DataFrame: Working With Information Efficiently
Source: https://realpython.com/pandas-dataframe/
Belum ada Komentar untuk "Use the Pandas Dataframe Dept stats Again in Python"
Posting Komentar